2.5.1 Tab-Delimited Text Format
OpenClinicas standard tabular (non-CDISC XML) data export formats are HTML, tab-delimited, Excel, and SPSS. The HTML, tab-delimited, and Excel formats each contain 2 tables a header table that contains reference information about the dataset contents, and the data table. The SPSS data export format has a data table similar in structure and format to the others, but does not have a header table. Instead it includes a separate .sps syntax file that describes the dataset. The following image shows a sample output for HTML format.
2.5.1.1 Header Table Format
The header table includes the following information:
- Dataset name
- Dataset description
- Study name
- Protocol ID the study protocol ID
- Date the date the data set was created
- Subjects the number of subject records in the dataset
- Study Event Definitions the number of study event definitions included in the dataset
- For each of the included study event definitions, the name of the event defintion plus an identifier which is used to reference the event definition in the data table
For each of the included case report forms (CRFs), the name of the CRF plus an identifier which is used to reference the CRF in the data table
2.5.1.2 Data Table Format
To avoid duplication and confusion amongst the data points collected in a study, certain identifiers and ordinal numbers must be appended to each variable name. These variable names can be used in multiple CRFs across multiple Events.
These appendages will help identify the event, CRF and item the value was collected in. The identifiers are defined in the header table for tab, HTML, and Excel formats. The identifiers are defined in a separate syntax (.sps) file for SPSS. The following scheme will be implemented:
E1 = E specifies that the appendage represents the event. 1 specifies which event the variable is from, as defined in the header table. If the event is repeating, it would be represented as E1_1, E1_2, E1_3 etc.
C1 = C specifies that the appendage represents a CRF. 1 specifies which CRF the variable is from, as defined in the header table
For repeating events and repeating groups, additional information must be provided to detail which occurrence of the event and/or which repeat of the group the item value comes from. This is done by appending _X where X is the ordinal or repeat number. As an example, an item called DEMO appearing in the 3rd occurrence of a repeating event, and the 5th repeat of the group called Example would be identified in the following way.
DEMO_E1_3_C1_5
For an item in a repeating event, but not part of a repeating group, the variable would be identified in the following way:
DEMO_E1_3_C1
2.5.1.3 Variable naming convention
To avoid duplication and confusion amongst the data points collected in a study, certain identifiers and ordinal numbers must be appended to each variable name. These variable names can be used in multiple CRFs across multiple Events.
These appendages will help identify the event, CRF and item the value was collected in. The identifiers are defined in the header table for tab, HTML, and Excel formats. The identifiers are defined in a separate syntax (.sps) file for SPSS. The following scheme will be implemented:
E1 = E specifies that the appendage represents the event. 1 specifies which event the variable is from, as defined in the header table. If the event is repeating, it would be represented as E1_1, E1_2, E1_3 etc.
C1 = C specifies that the appendage represents a CRF. 1 specifies which CRF the variable is from, as defined in the header table
For repeating events and repeating groups, additional information must be provided to detail which occurrence of the event and/or which repeat of the group the item value comes from. This is done by appending _X where X is the ordinal or repeat number. As an example, an item called DEMO appearing in the 3rd occurrence of a repeating event, and the 5th repeat of the group called Example would be identified in the following way.
DEMO_E1_3_C1_5
For an item in a repeating event, but not part of a repeating group, the variable would be identified in the following way:
DEMO_E1_3_C1
2.5.2 HTML Format
You can download and view a dataset file in HTML format (.html file).
Example of Dataset File in HTML Format (Partial View):
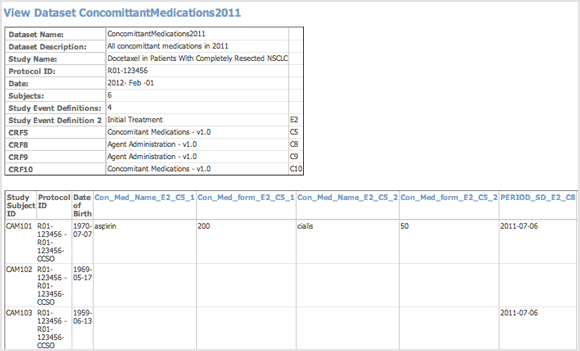
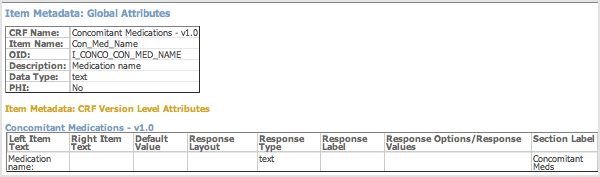
When viewing the HTML file, you can view the metadata for an Item by clicking its column header.
Example of Item Metadata for HTML Dataset File (Partial View):
2.5.3 Excel Spreadsheet Format
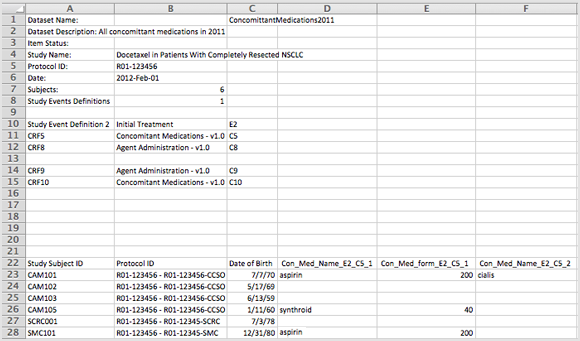
When you download to the Excel Spreadsheet Format, the dataset is an .xls file.
Example of Dataset File in Excel Spreadsheet Format (Partial View):
2.5.4 SPSS Format
When you select the SPSS format, the extracted .zip file contains two different files: a .dat file, which is a tab-delimited data file, and an .sps file, which is an SPSS data definition script.
To access the data, save the .dat and .sps files to the same location, then open the .sps file in the IBM SPSS program. If the .sps and .dat files are not in the same location, change the FILE location in the .sps file to point to the physical location of the .dat file. Then from SPSS, select Run > All to load the data into the application.
You can preview the .dat file by opening it in a text editor.
See SPSS Syntax File Specifications for more information.
2.5.5 Data Mart Format
Select the Data Mart format for use with the OpenClinica Enterprise Edition. The exported dataset is an .sql file. Use the file with the Data Mart feature to insert the data into a database.
See Data Mart for more information.
2.5.6 CDISC ODM Formats
When you select one of the CDISC ODM formats for the dataset, OpenClinica exports the dataset to an .xml file that complies with the Operational Data Model (ODM) of the Clinical Data Interchange Standards Consortium (CDISC) standard. These are the different parameters for the available ODM formats:
- 1.3 or 1.2: refers to the version of the ODM specification.
- With extensions: Includes OpenClinica entities that are not part of the ODM specification, such as the Subject Group Class and its attributes.
- Full: Includes Discrepancy Notes and the Audit Log.
CDISC ODM is a vendor neutral, platform independent format for interchange and archive of data collected in clinical trials. The model represents study metadata, data, and administrative data associated with a clinical trial. The ODM has been designed to be compliant with guidance and regulations published by the FDA for computer systems used in clinical trials.
The ODM model categorizes a clinical study’s data into several kinds of entities including subjects, study events, forms, item groups, items, and annotations. The metadata of a study describes the types of study events, forms, item groups, and items that are allowed in the study. The clinical data of a study will typically have many actual entities corresponding to their definitions described in the metadata.
Like any XML file, an ODM file consists of a tree of elements that correspond to entities. Each element consists of required attributes and optional attributes. An ODM file type must be either Snapshot or Transactional. A Snapshot file shows the current state of the included data. A Transactional file shows both latest state and (optionally) some prior states of an included entity. An ODM file has a Granularity attribute which describes the coverage information of the ODM file.
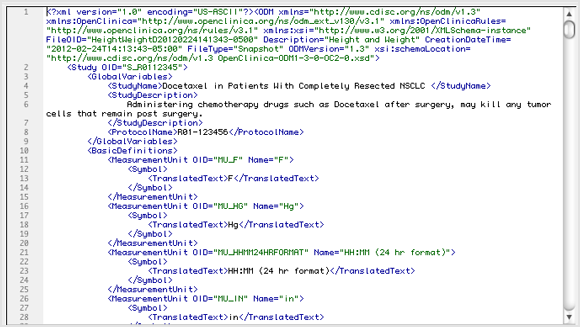
The ODM file consists of two parts: metadata followed by Subject data. The metadata provides OIDs for the Study, units (as defined when the CRFs were created), Event information, CRF information including Item Groups and Items with information about validations, and user account information. The Subject data provides Subject information, Event information, CRF information, and then the values.
CDISC ODM Format XML File – Metadata Section (Partial View):
CDISC ODM Format XML File – Subject Data Section (Partial View):

For more details about the ODM specification and its use in OpenClinica, see CDISC ODM Representation in OpenClinica in the OpenClinica Technical Documentation, and the ODM Final Version 1.3 for Implementation at the CDISC web site.
2.5.7 SAS Data and Syntax
OpenClinica version 3.11 introduced the SAS Data and Syntax extract format, which were tested using SAS Studio. This extract format functions as follows:
- The output includes three files:
- SAS_DATA.xml – The extracted data.
- SAS_Format.sas – For items defined as single-select or radio button, OpenClinica creates the library and maps response values to the appropriate response text.
Note: Because multi-select and checkbox items include multiple values in a string format in OpenClinica (e.g., 1,2,7), these cannot be mapped to individual response text options.
- SAS_MAP.xml – A mapping file that maps the data to the appropriate structures (e.g., LIBNAME, Table, Column) OpenClinica forces appropriate object names as required by SAS. For example, all Studies start with “S” and all Table names and Column names start with an underscore.
- Once the extract files are downloaded, upload the SAS_DATA and SAS_MAP files into SAS Studio.
- Open the SAS_Format.sas file, copy the text, and paste it into SAS Studio.
- Click the Run icon.
- This generates all the data tables based on Item Groups.
- OpenClinica Items become SAS Column Names.
- Tables include the master set of items (i.e., Item Groups span CRF Versions, though the SAS file does not indicate which version of the CRF was the source for the item.)
- There are two resulting data types: Numeric or Char. All OpenClinica items that are Integer or Real are classified as Numeric. All other OpenClinica data types are classified as Char.
- The SAS datasets/tables are generated from the OpenClinica metadata. Tables are created for all Item Groups in the extract. If no data was entered for a specific item group, the SAS table is still created, but is empty.
The following apply due to SAS name limitations:
- OpenClinica and DataMart allow 3,999 single-byte characters in a text field. When this size string is extracted to SAS, the full string is in the SAS_DATA.xml file.
- SAS data set names must not exceed 32 characters and must start with either a letter (A-Z) or underscore. As a result, Openclinica uses a modified Item Group OID for the data set name as follows:
- If group is Ungrouped use the CRF Name, otherwise:
- To reduce the number of characters the pre-pended IG is removed (This means Group labels start with “_” + 5CHAR (of CRF Name) + _GROUPLABEL)
- If the resulting value exceeds 35 characters, OpenClinica appends the dataset name with the three- or four-digit number appended to the IG_OID
- SAS column names must not exceed 32 characters and must start with a letter (A-Z) or underscore. As a result, OpenClinica uses a modified Item OID for the column names as follows:
- Truncate from the left to remove the I_5CHAR prefix to each Item Name.
- Use the portion of the OID starting with _ (underscore) followed by ITEMNAME (this ensures no Column Names start with a number.)
- Retain appended three- or four-digit numbers to ensure item/column name uniqueness.
